CLOCK/Salmonella Dataset
Table of Contents
- 1 Network Key
- 2 Cluster 1
- 2.1 TFs included in Network
- 2.2 Networks
- 2.2.1 TF network (CLOCKTRIMALL)
- 2.2.2 Fully-connected TF network (CLOCKTRIMTFALL)
- 2.2.3 Full network (CLOCKTRIM)
- 2.2.4 Proteins significant in WT infected vs uninfected day (CLOCKTRIM0)
- 2.2.5 Proteins significant in WT infected vs uninfected night (CLOCKTRIM1)
- 2.2.6 Proteins significant in CLOCK mutant infected vs uninfected day (CLOCKTRIM2)
- 2.2.7 Proteins significant in CLOCK mutant infected vs uninfected night (CLOCKTRIM3)
- 2.2.8 Difference between CLOCK mutant and WT day (CLOCKTRIM0REMOVED)
- 2.2.9 Difference between CLOCK mutant and WT night (CLOCKTRIM1REMOVED)
- 2.3 GO analysis (Top 20 enriched annotations)
- 3 Cluster 2
- 3.1 Normal
- 3.2 TFs
- 3.3 All
- 3.4 Significant in WT infected vs uninfected day
- 3.5 Significant in WT infected vs uninfected night
- 3.6 Significant in CLOCK mutant infected vs uninfected day
- 3.7 Significant in CLOCK mutant infected vs uninfected night
- 3.8 Difference between CLOCK mutant and WT day
- 3.9 Difference between CLOCK mutant and WT night
- 4 Cluster 3
- 4.1 Normal
- 4.2 TFs
- 4.3 All
- 4.4 Significant in WT infected vs uninfected day
- 4.5 Significant in WT infected vs uninfected night
- 4.6 Significant in CLOCK mutant infected vs uninfected day
- 4.7 Significant in CLOCK mutant infected vs uninfected night
- 4.8 Difference between CLOCK mutant and WT day
- 4.9 Difference between CLOCK mutant and WT night
- 5 Cluster 4
- 5.1 Normal
- 5.2 TFs
- 5.3 All
- 5.4 Significant in WT infected vs uninfected day
- 5.5 Significant in WT infected vs uninfected night
- 5.6 Significant in CLOCK mutant infected vs uninfected day
- 5.7 Significant in CLOCK mutant infected vs uninfected night
- 5.8 Difference between CLOCK mutant and WT day
- 5.9 Difference between CLOCK mutant and WT night
1 Network Key
If node contains this color then it is significant at .05 p-value in that dataset. Can be significant in multiple, or none.
- Blue = WT infected vs uninfected day
- Green = WT infected vs uninfected night
- Brown = CLOCK mutant infected vs uninfected day
- Orange = CLOCK mutant infected vs uninfected night
2 Cluster 1
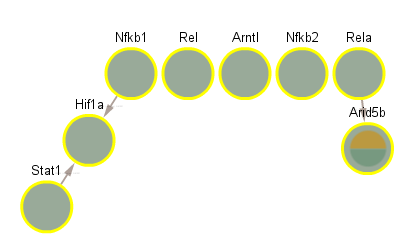
2.1 TFs included in Network
Nfkb2
{'accessions': ['Q9WTK5'],
'comment_similarity': ['Contains 7 ANK repeats.',
'Contains 1 death domain.',
'Contains 1 RHD (Rel-like) domain.'],
'name': 'Nfkb2',
'recommendedName_fullName': ['Nuclear factor NF-kappa-B p100 subunit']}
Hif1a
{'accessions': ['Q61221',
'O08741',
'O08993',
'Q61664',
'Q61665',
'Q8C681',
'Q8CC19',
'Q8CCB6',
'Q8R385',
'Q9CYA8'],
'comment_similarity': ['Contains 1 basic helix-loop-helix (bHLH) domain.',
'Contains 1 PAC (PAS-associated C-terminal) domain.',
'Contains 2 PAS (PER-ARNT-SIM) domains.'],
'name': 'Hif1a',
'recommendedName_fullName': ['Hypoxia-inducible factor 1-alpha'],
'recommendedName_shortName': ['HIF-1-alpha', 'HIF1-alpha']}
Ovol1
{'accessions': ['Q9WTJ2', 'Q545S2'],
'comment_similarity': ['Contains 4 C2H2-type zinc fingers.'],
'name': 'Ovol1',
'recommendedName_fullName': ['Putative transcription factor Ovo-like 1'],
'recommendedName_shortName': ['mOvo1', 'mOvo1a']}
Prdm1
{'accessions': ['Q60636', 'A2VDE8', 'Q3UET9'],
'comment_similarity': ['Contains 4 C2H2-type zinc fingers.',
'Contains 1 SET domain.'],
'name': 'Prdm1',
'recommendedName_fullName': ['PR domain zinc finger protein 1']}
Smad2
{'accessions': ['Q62432', 'Q6GU18', 'Q6VP00', 'Q9D8P6'],
'comment_similarity': ['Belongs to the dwarfin/SMAD family.',
'Contains 1 MH1 (MAD homology 1) domain.',
'Contains 1 MH2 (MAD homology 2) domain.'],
'name': 'Smad2',
'recommendedName_fullName': ['Mothers against decapentaplegic homolog 2'],
'recommendedName_shortName': ['MAD homolog 2',
'Mothers against DPP homolog 2']}
Stat3
{'accessions': ['P42227', 'A2A5D1', 'B7ZC17'],
'comment_similarity': ['Belongs to the transcription factor STAT family.',
'Contains 1 SH2 domain.'],
'name': 'Stat3',
'recommendedName_fullName': ['Signal transducer and activator of transcription 3']}
Arntl/CLOCK
{'accessions': ['Q9WTL8', 'O88295', 'Q921S4', 'Q9R0U2', 'Q9WTL9'],
'comment_similarity': ['Contains 1 basic helix-loop-helix (bHLH) domain.',
'Contains 1 PAC (PAS-associated C-terminal) domain.',
'Contains 2 PAS (PER-ARNT-SIM) domains.'],
'name': 'Arntl',
'recommendedName_fullName': ['Aryl hydrocarbon receptor nuclear translocator-like protein 1']}
Nfkb1
{'accessions': ['P25799',
'Q3TZE8',
'Q3V2V6',
'Q6TDG8',
'Q75ZL1',
'Q80Y21',
'Q8C712'],
'comment_similarity': ['Contains 7 ANK repeats.',
'Contains 1 death domain.',
'Contains 1 RHD (Rel-like) domain.'],
'name': 'Nfkb1',
'recommendedName_fullName': ['Nuclear factor NF-kappa-B p105 subunit']}
Irf8
{'accessions': ['P23611'],
'comment_similarity': ['Belongs to the IRF family.',
'Contains 1 IRF tryptophan pentad repeat DNA-binding domain.'],
'name': 'Irf8',
'recommendedName_fullName': ['Interferon regulatory factor 8'],
'recommendedName_shortName': ['IRF-8']}
Rel
{'accessions': ['P15307'],
'comment_similarity': ['Contains 1 RHD (Rel-like) domain.'],
'name': 'Rel',
'recommendedName_fullName': ['Proto-oncogene c-Rel']}
Rela
{'accessions': ['Q04207', 'Q62025'],
'comment_similarity': ['Contains 1 RHD (Rel-like) domain.'],
'name': 'Rela',
'recommendedName_fullName': ['Transcription factor p65']}
Myc
{'accessions': ['P01108', 'P70247', 'Q3UM70', 'Q61422'],
'comment_similarity': ['Contains 1 basic helix-loop-helix (bHLH) domain.'],
'name': 'Myc',
'recommendedName_fullName': ['Myc proto-oncogene protein']}
Hnf4g
{'accessions': ['Q9WUU6'],
'comment_similarity': ['Belongs to the nuclear hormone receptor family. NR2 subfamily.',
'Contains 1 nuclear receptor DNA-binding domain.'],
'name': 'Hnf4g',
'recommendedName_fullName': ['Hepatocyte nuclear factor 4-gamma'],
'recommendedName_shortName': ['HNF-4-gamma']}
Atf1
{'accessions': ['P81269'],
'comment_similarity': ['Belongs to the bZIP family. ATF subfamily.',
'Contains 1 bZIP domain.',
'Contains 1 KID (kinase-inducible) domain.'],
'name': 'Atf1',
'recommendedName_fullName': ['Cyclic AMP-dependent transcription factor ATF-1'],
'recommendedName_shortName': ['cAMP-dependent transcription factor ATF-1']}
2.2 Networks
2.2.1 TF network (CLOCKTRIMALL)
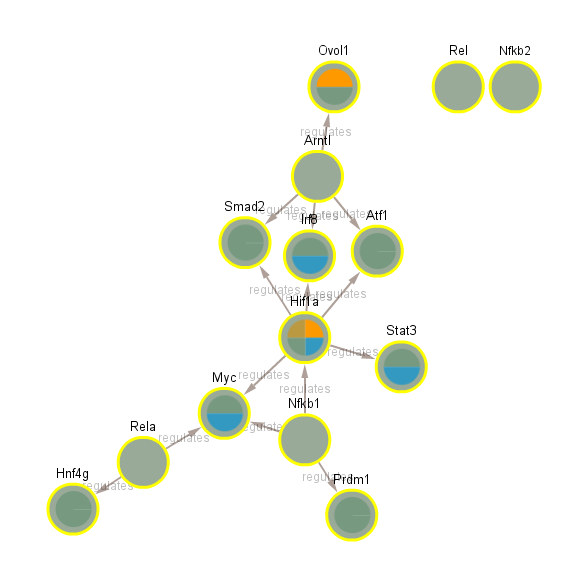
2.2.2 Fully-connected TF network (CLOCKTRIMTFALL)

2.2.3 Full network (CLOCKTRIM)
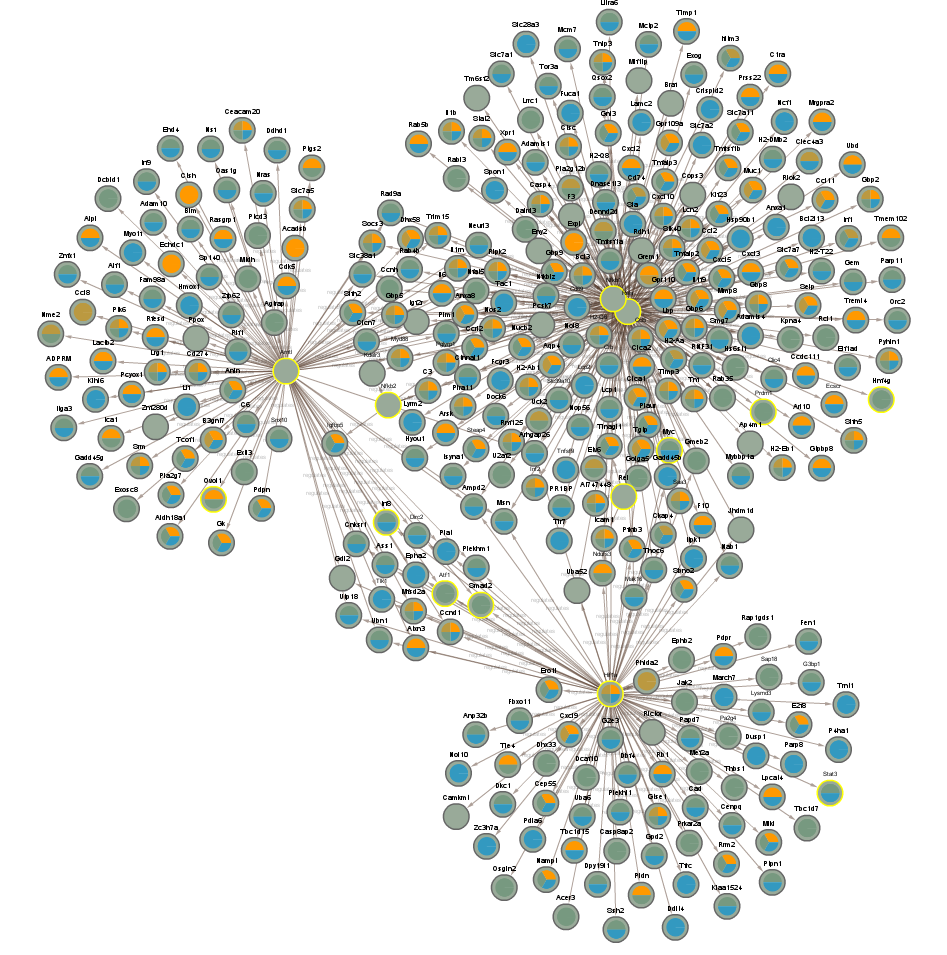
2.2.4 Proteins significant in WT infected vs uninfected day (CLOCKTRIM0)
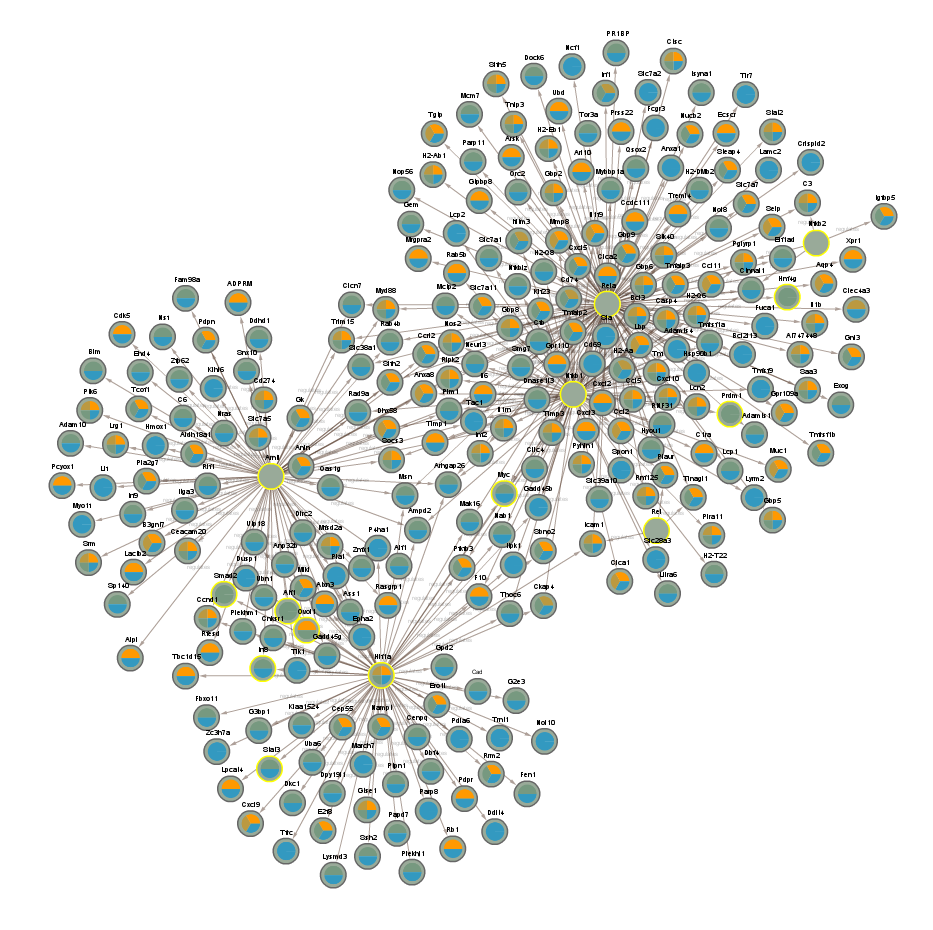
2.2.5 Proteins significant in WT infected vs uninfected night (CLOCKTRIM1)
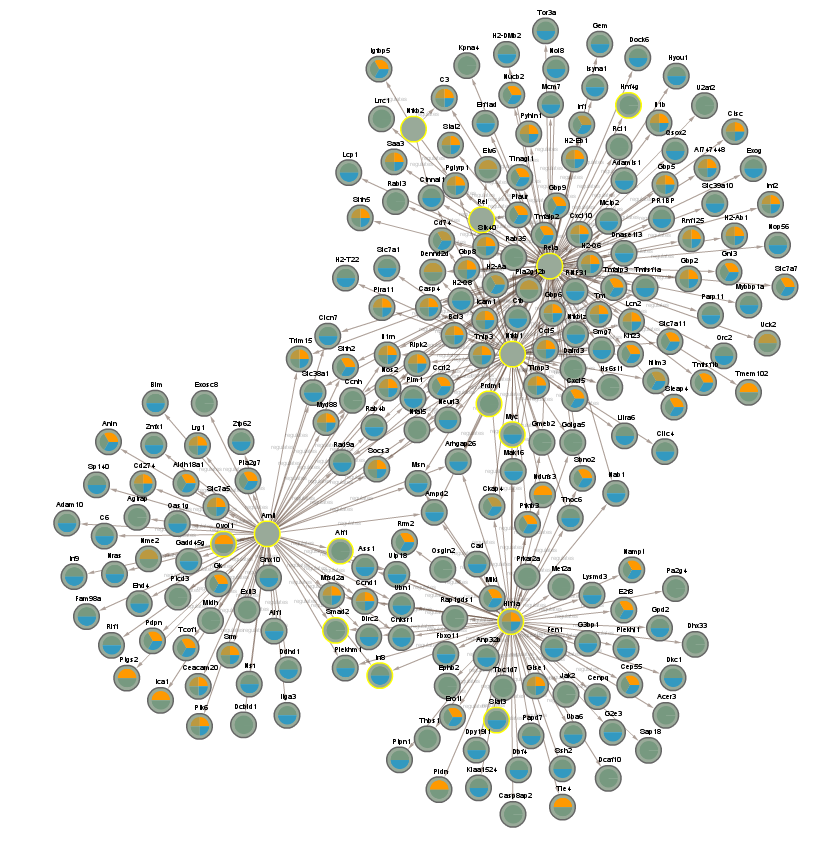
2.2.6 Proteins significant in CLOCK mutant infected vs uninfected day (CLOCKTRIM2)

2.2.7 Proteins significant in CLOCK mutant infected vs uninfected night (CLOCKTRIM3)
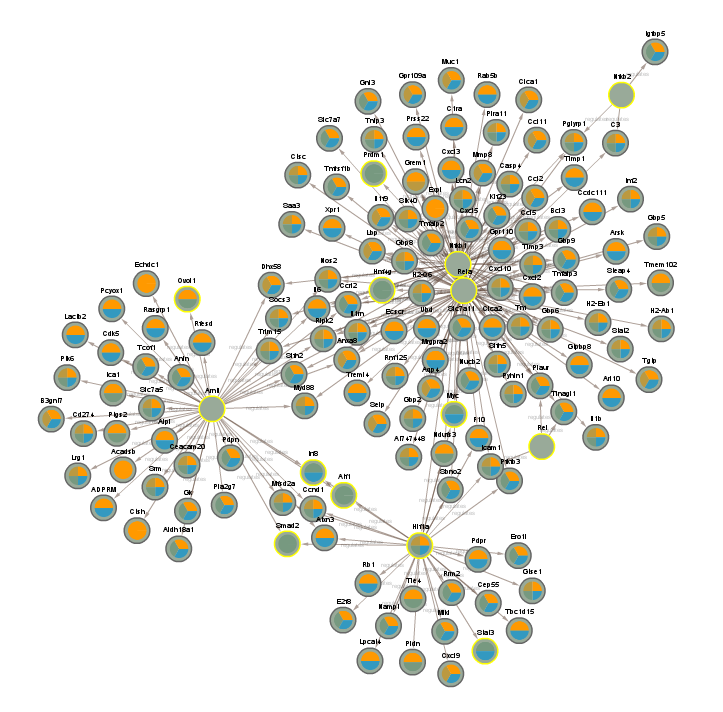
2.2.8 Difference between CLOCK mutant and WT day (CLOCKTRIM0REMOVED)

2.2.9 Difference between CLOCK mutant and WT night (CLOCKTRIM1REMOVED)
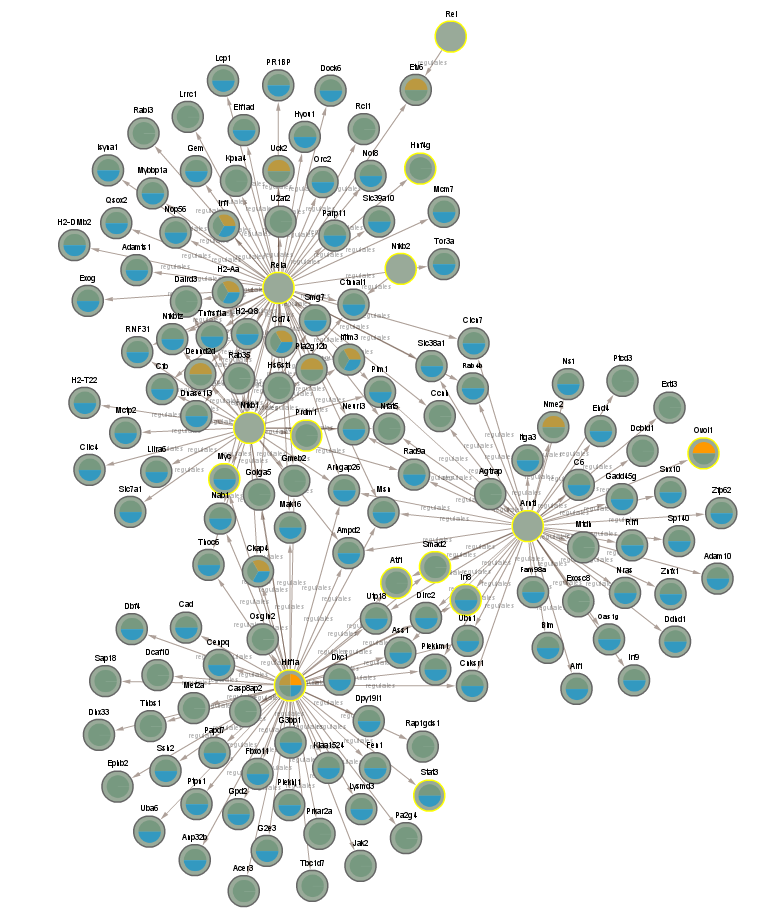
2.3 GO analysis (Top 20 enriched annotations)
| GO Term | Count | Percentage | p-value | Fold Enrichment | Benjamini p-value | |
|---|---|---|---|---|---|---|
| 1 | immune response | 53 | 15.8682634730539 | 1.45855715729538e-24 | 5.62136255776196 | 2.5568506967388e-21 |
| 2 | response to wounding | 39 | 11.6766467065868 | 6.00863372959416e-18 | 5.61463807424987 | 5.26656746398928e-15 |
| 3 | inflammatory response | 32 | 9.5808383233533 | 1.20226439707048e-17 | 7.10483660130719 | 7.02523162688182e-15 |
| 4 | defense response | 42 | 12.5748502994012 | 1.65261964360520e-16 | 4.68336397058824 | 4.86277684785819e-14 |
| 5 | mmu04621:NOD-like receptor signaling pathway | 13 | 3.89221556886228 | 1.07871580920171e-08 | 9.04608294930875 | 1.26209670292532e-06 |
| 6 | inflammatory response | 13 | 3.89221556886228 | 1.21498784480603e-08 | 9.35744234800839 | 3.85150407078427e-06 |
| 7 | cytokine | 18 | 5.38922155688623 | 1.81339374759809e-08 | 5.70962583946274 | 2.87422498335133e-06 |
| 8 | PIRSF005552:guanine nucleotide-binding protein 1 | 6 | 1.79640718562874 | 1.88446452613451e-07 | 36.1065088757396 | 3.65579470036348e-05 |
| 9 | IPR003191:Guanylate-binding protein, C-terminal | 6 | 1.79640718562874 | 2.01971601551270e-07 | 37.4746835443038 | 0.000150861420868398 |
| 10 | IPR001811:Small chemokine, interleukin-8-like | 9 | 2.69461077844311 | 2.20869685477391e-07 | 13.6731953472460 | 8.24914340129546e-05 |
| 11 | cytokine activity | 17 | 5.08982035928144 | 3.94942817405200e-07 | 4.88318201469952 | 0.000178893128349955 |
| 12 | chemokine activity | 9 | 2.69461077844311 | 5.04559487901997e-07 | 12.2457505631784 | 0.000114276222805798 |
| 13 | chemokine receptor binding | 9 | 2.69461077844311 | 6.24279723051955e-07 | 11.9317569589943 | 9.42618246763116e-05 |
| 14 | regulation of cytokine production | 15 | 4.49101796407186 | 7.1297835304467e-07 | 5.39092255607279 | 0.000249939059709492 |
| 15 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 12 | 3.59281437125748 | 8.30188111897995e-07 | 7.13655462184874 | 0.000242523980352738 |
| 16 | adaptive immune response | 12 | 3.59281437125748 | 8.30188111897995e-07 | 7.13655462184874 | 0.000242523980352738 |
| 17 | SM00199:SCY | 9 | 2.69461077844311 | 9.92421443649605e-07 | 11.1052702702703 | 0.000168697498120540 |
| 18 | immune effector process | 14 | 4.19161676646707 | 1.35897526105141e-06 | 5.55065359477124 | 0.000340268560009682 |
| 19 | IPR015894:Guanylate-binding protein, N-terminal | 6 | 1.79640718562874 | 1.94578279769702e-06 | 25.944011684518 | 0.000484383036624569 |
| 20 | extracellular region part | 34 | 10.1796407185629 | 2.10414239539143e-06 | 2.47419512535792 | 0.000506970297911691 |
3 Cluster 2
3.1 Normal

3.2 TFs
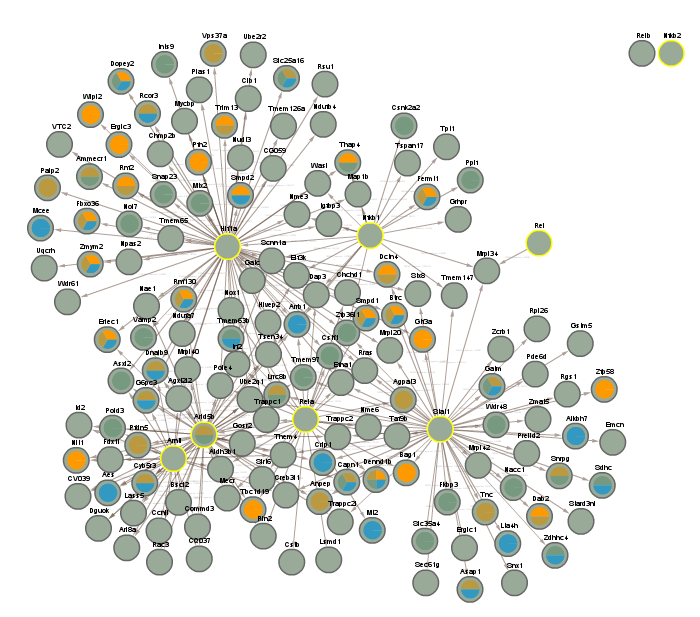
3.3 All
3.4 Significant in WT infected vs uninfected day
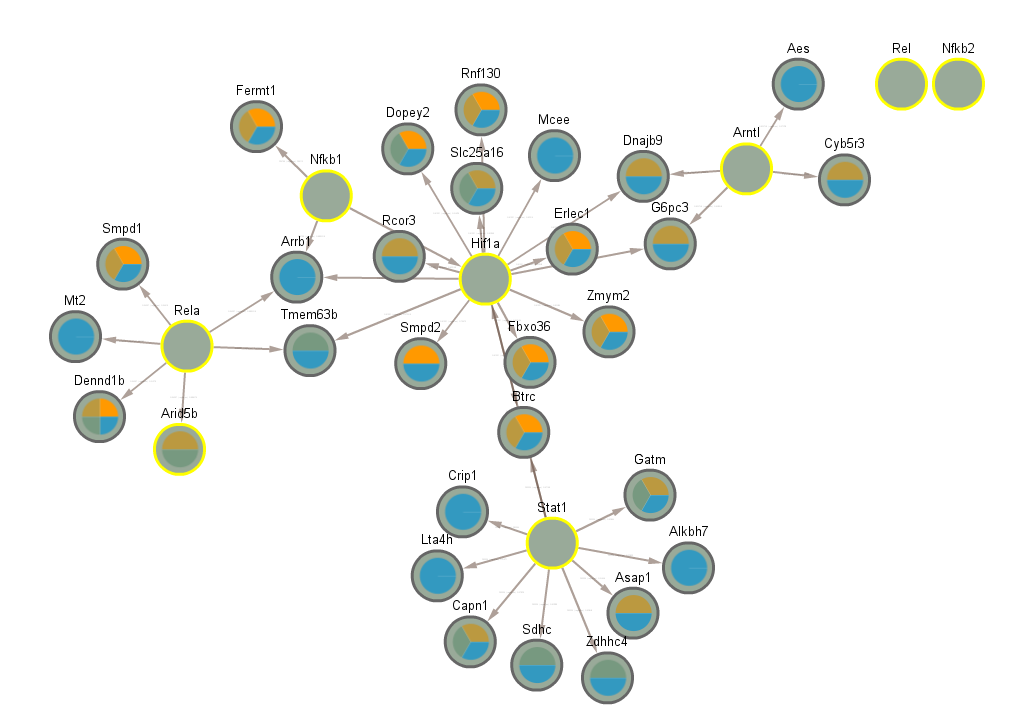
3.5 Significant in WT infected vs uninfected night
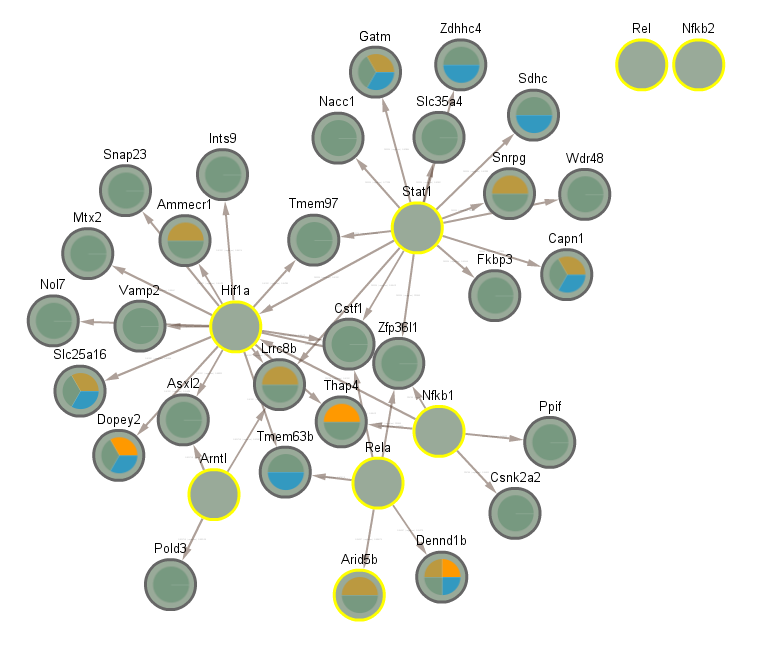
3.6 Significant in CLOCK mutant infected vs uninfected day
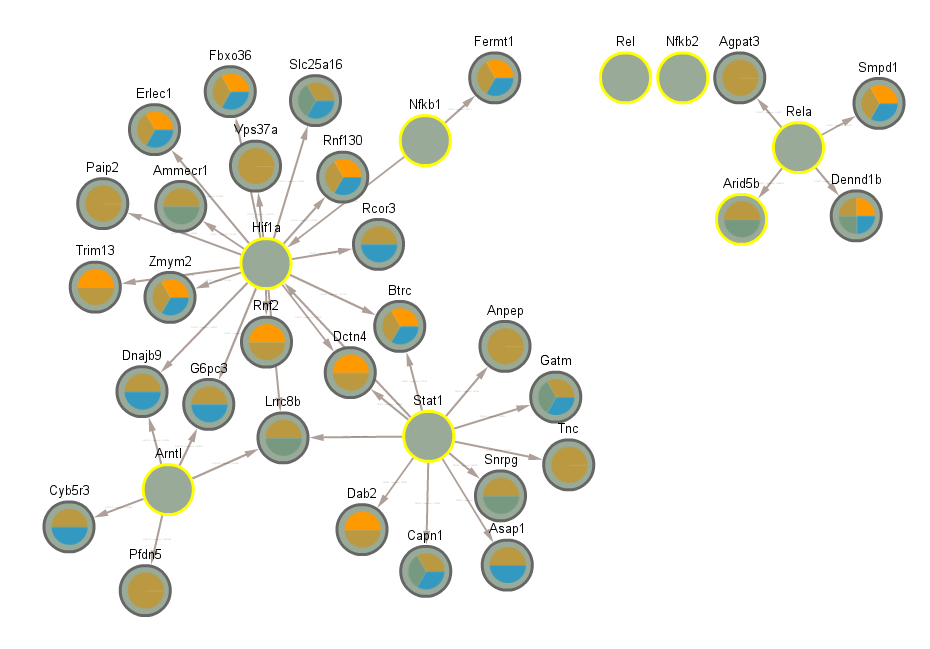
3.7 Significant in CLOCK mutant infected vs uninfected night
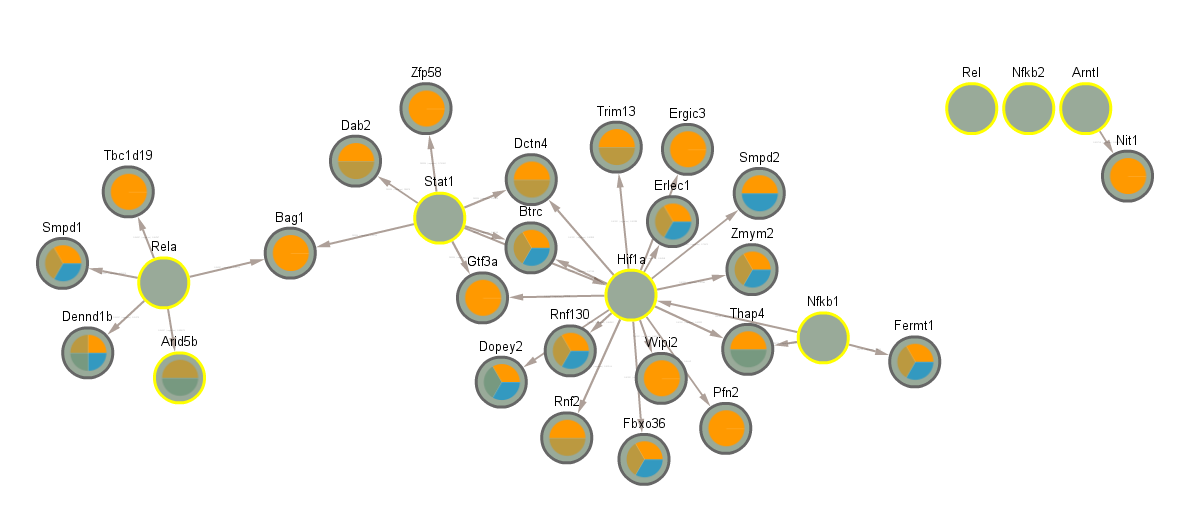
3.8 Difference between CLOCK mutant and WT day

3.9 Difference between CLOCK mutant and WT night
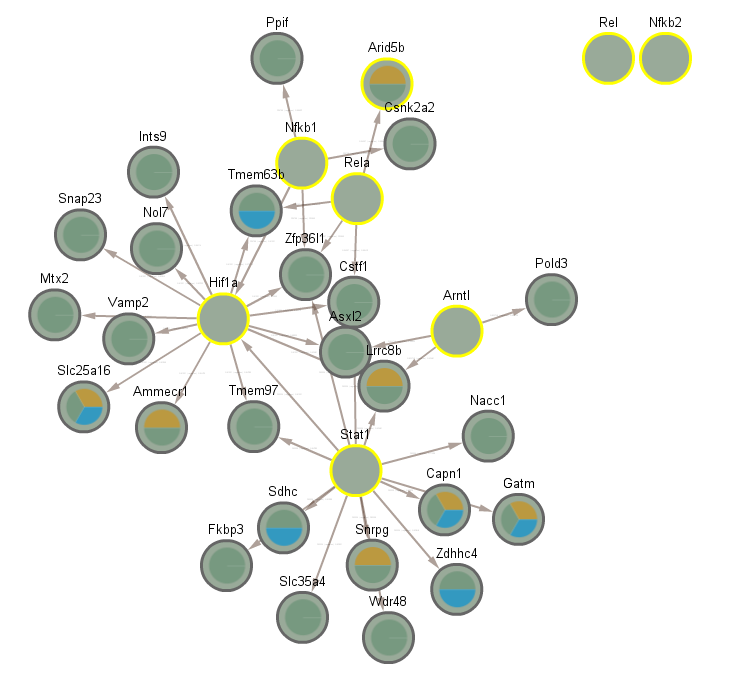
4 Cluster 3
4.1 Normal
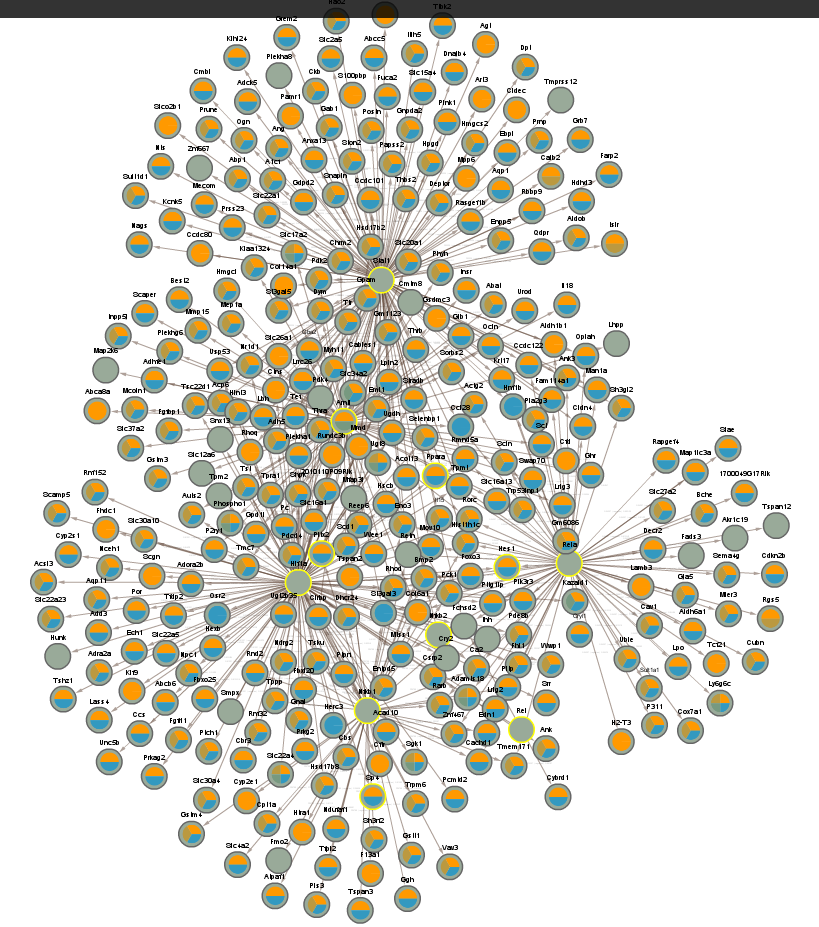
4.2 TFs
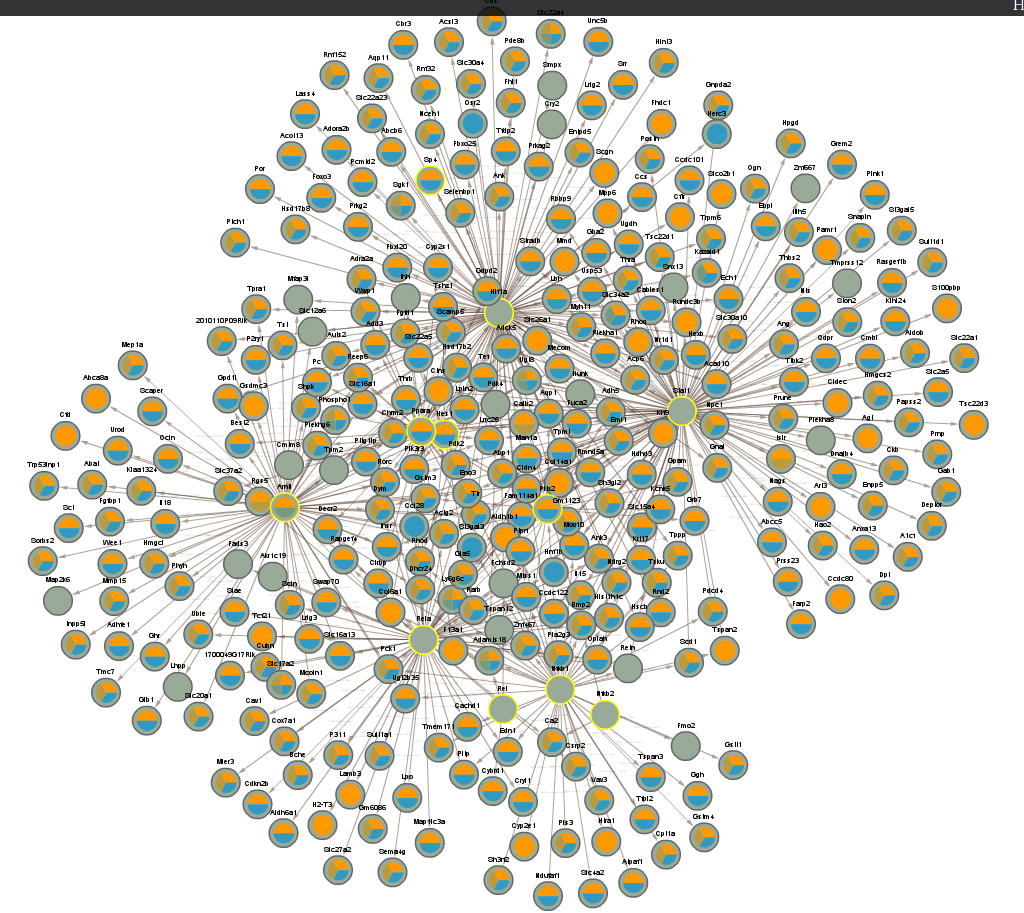
4.3 All

4.4 Significant in WT infected vs uninfected day
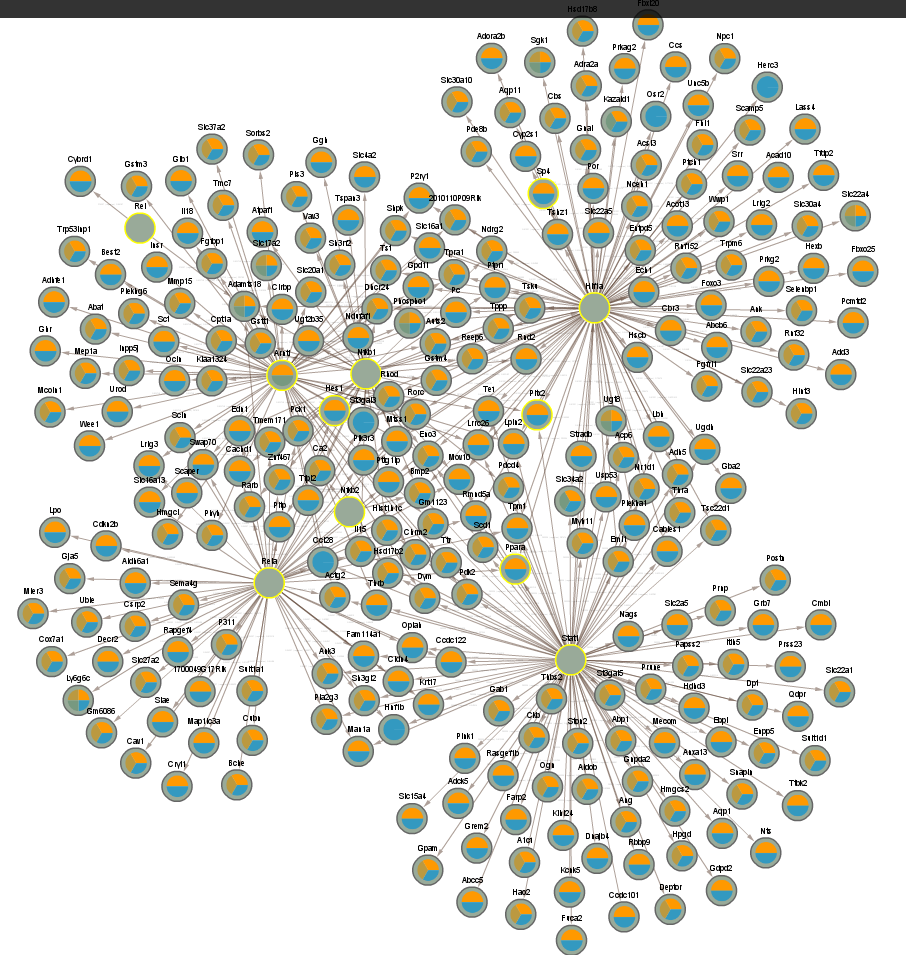
4.5 Significant in WT infected vs uninfected night
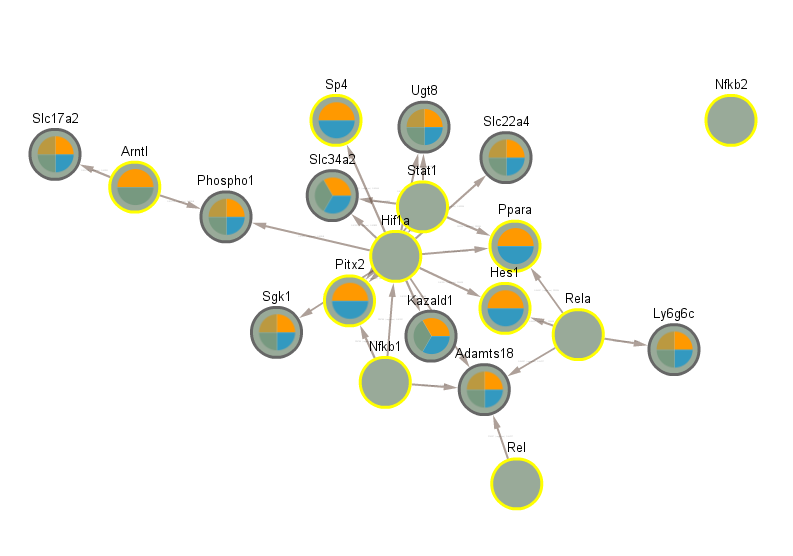
4.6 Significant in CLOCK mutant infected vs uninfected day
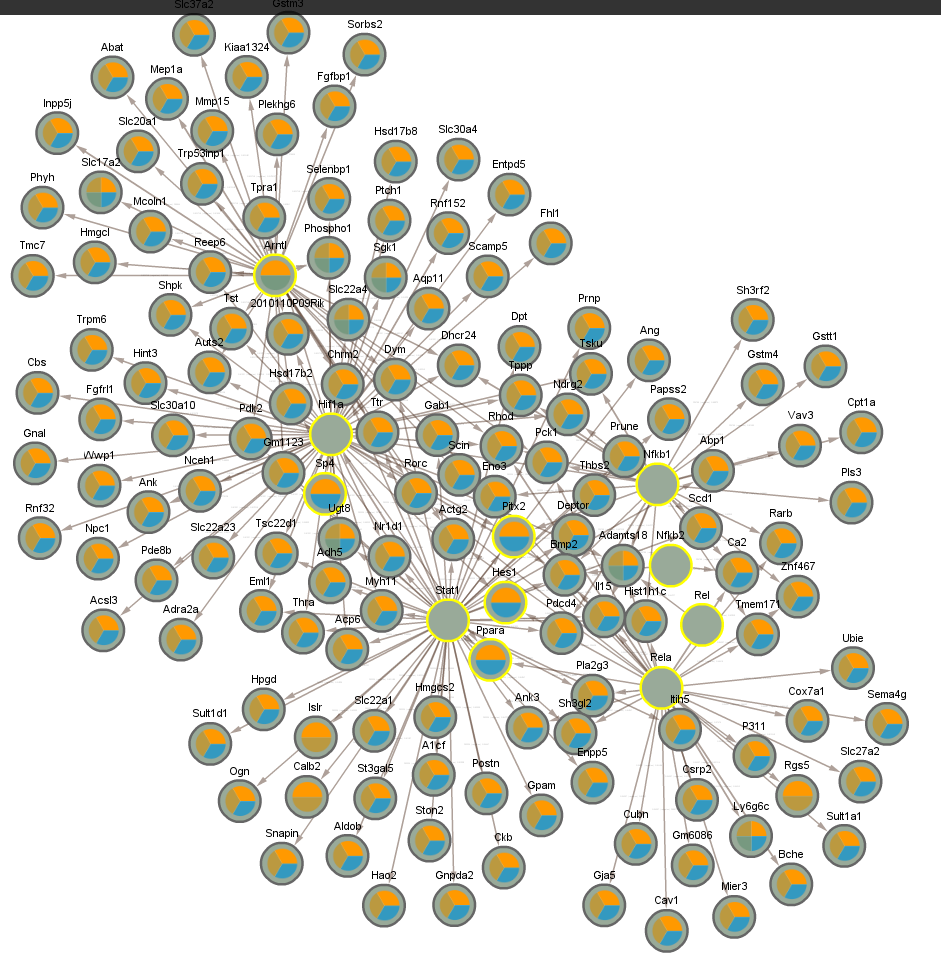
4.7 Significant in CLOCK mutant infected vs uninfected night
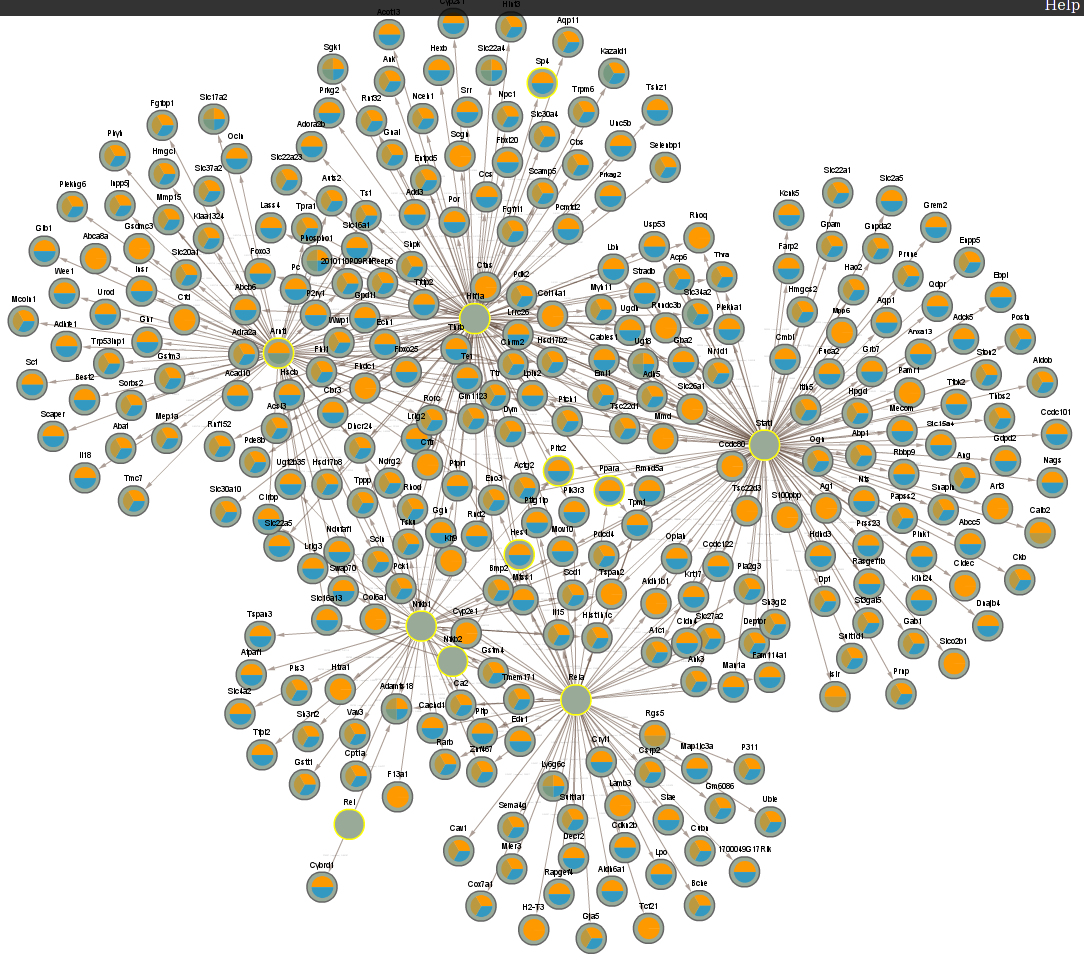
4.8 Difference between CLOCK mutant and WT day
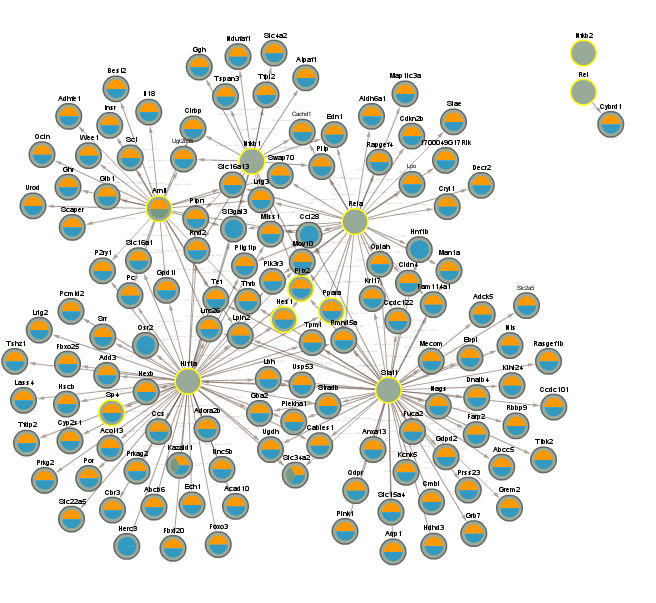
4.9 Difference between CLOCK mutant and WT night
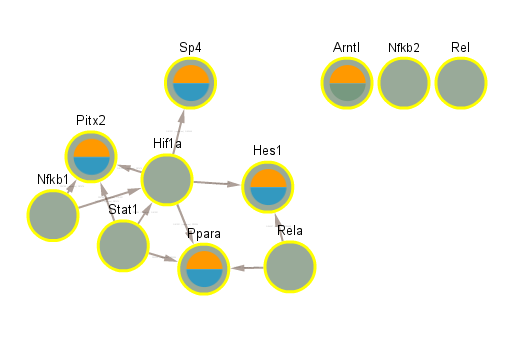
5 Cluster 4
5.1 Normal
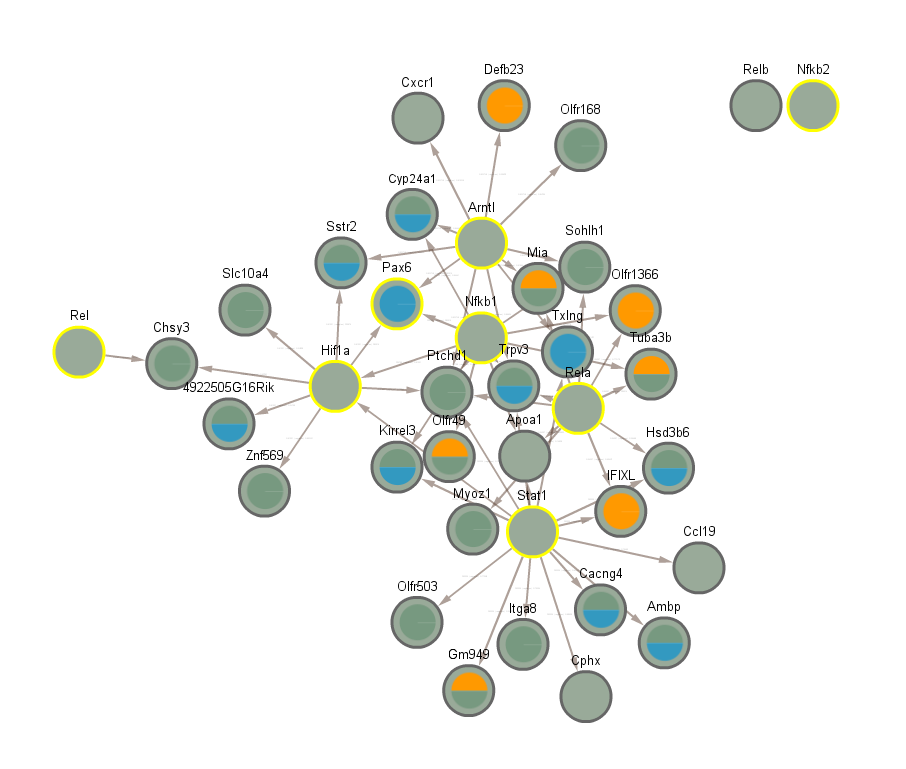
5.2 TFs
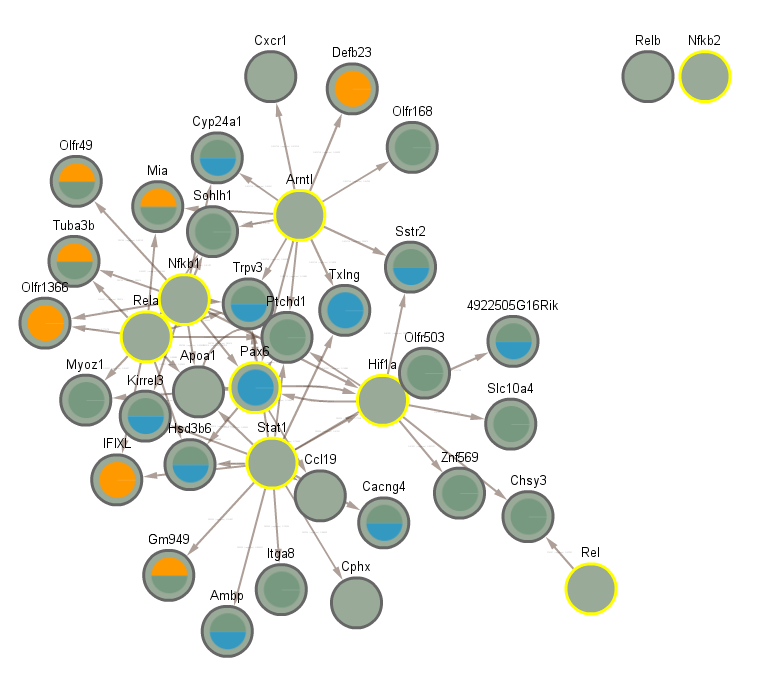
5.3 All
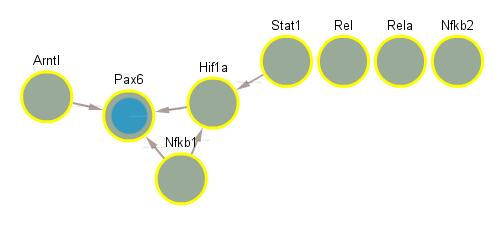
5.4 Significant in WT infected vs uninfected day

5.5 Significant in WT infected vs uninfected night
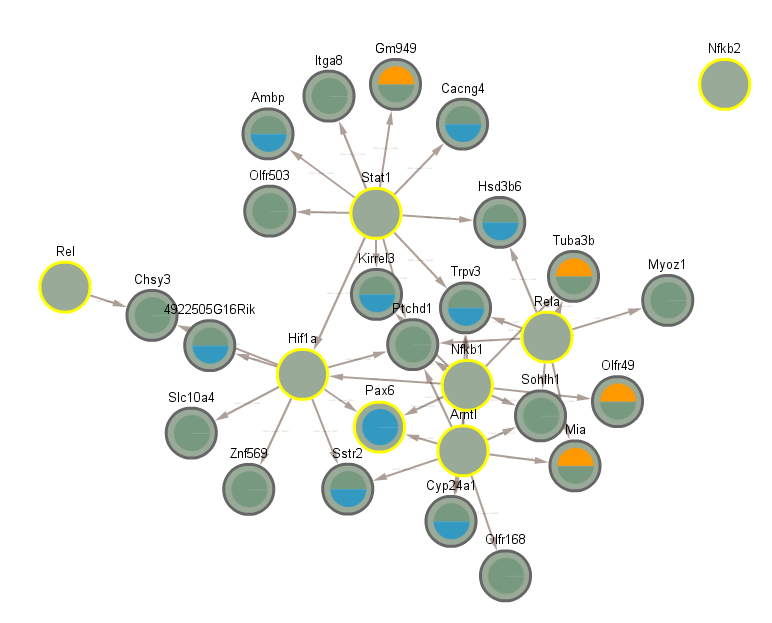
5.6 Significant in CLOCK mutant infected vs uninfected day
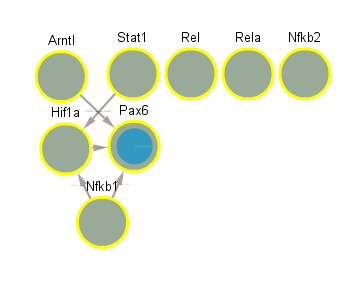
5.7 Significant in CLOCK mutant infected vs uninfected night

5.8 Difference between CLOCK mutant and WT day
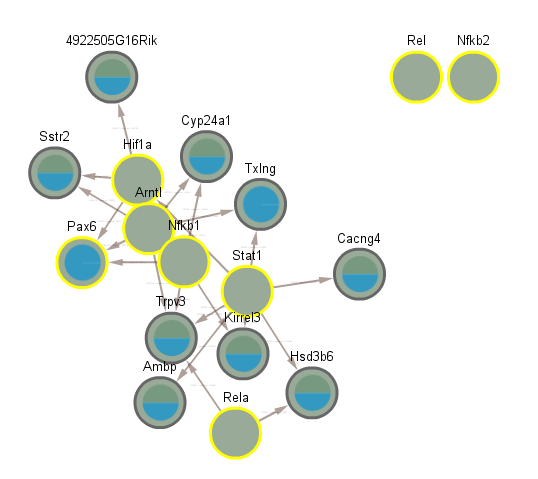
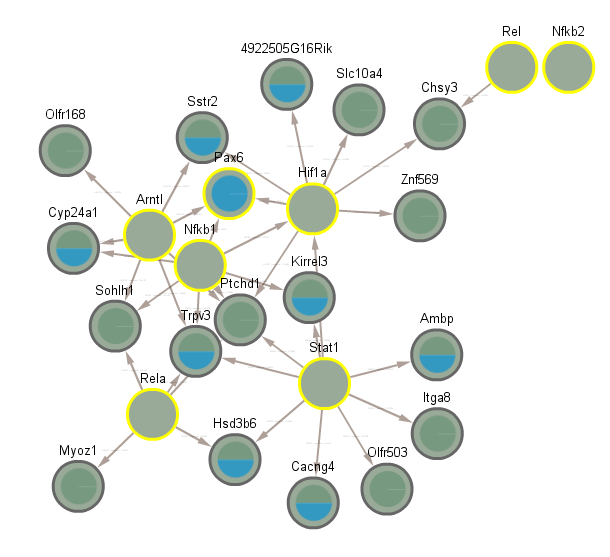
5.9 Difference between CLOCK mutant and WT night
Date: 2011-10-25 12:29:51 PDT
HTML generated by org-mode 6.30trans in emacs 23