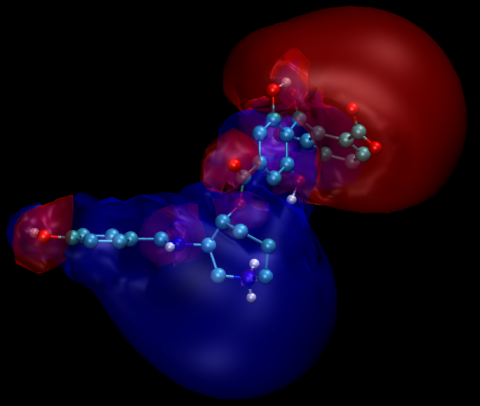
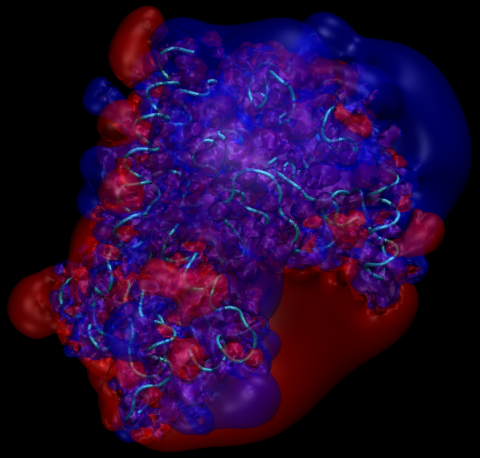
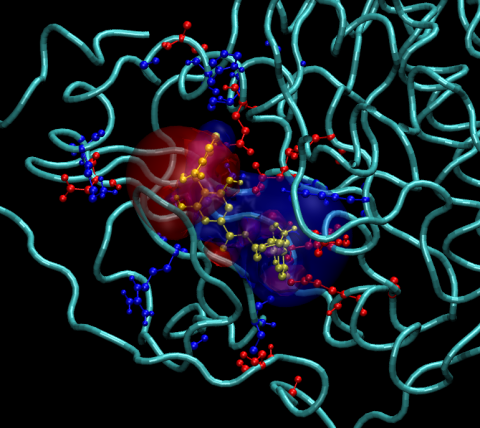
Visualization of the electrostatic potentials
This section of the example illustrates the calculation of lower-resolution electrostatic potentials suitable for interactive display and manipulation. First, we need to calculate electrostatic potentials for the three molecules. This is done with the following APBS input file which can be downloaded here.
Example 1. PKA/balanol/complex visualization APBS input
read
mol pqr bx6_7_lig_apbs.pqr # Read balanol (mol 1)
mol pqr bx6_7_apo_apbs.pqr # Read PKA (mol 2)
mol pqr bx6_7_bin_apbs.pqr # Read complex (mol 3)
end
elec name bal
mg-auto # Use the multigrid method
dime 97 97 97 # Grid dimensions
fglen 70 70 70 # Grid length
fgcent mol 3 # Center on complex
cglen 80 80 80 # Grid length
cgcent mol 3 # Center on complex
mol 1
lpbe
bcfl sdh # Monopole boundary conditions
ion 1 0.000 2.0 # Zero ionic strength
ion -1 0.000 2.0
pdie 2.0 # Solute dielectric
sdie 78.00 # Solvent dielectric
chgm spl0 # Charge disc method (linear)
srfm smol # Smoothed molecular surface
srad 0.0 # Solvent radius
swin 0.3 # Surface cubic spline window
sdens 10.0 # Sphere density
temp 298.15 # Temperature
gamma 0.105 # Surface tension (in kJ/mol/A^2)
calcenergy no
calcforce no
write pot dx ligand # Write potential to ligand.dx
end
elec name pka
mg-auto
dime 97 97 97
fglen 70 70 70
fgcent mol 3
cglen 80 80 80
cgcent mol 3
mol 2
lpbe
bcfl sdh
ion 1 0.000 2.0
ion -1 0.000 2.0
pdie 2.0
sdie 78.00
chgm spl0
srfm smol
srad 0.0
swin 0.3
sdens 10.0
temp 298.15
gamma 0.105
calcenergy no
calcforce no
write pot dx apo
end
elec name complex
mg-auto
dime 97 97 97
fglen 70 70 70
fgcent mol 3
cglen 80 80 80
cgcent mol 3
mol 3
lpbe
bcfl sdh
ion 1 0.000 2.0
ion -1 0.000 2.0
pdie 2.0
sdie 78.00
chgm spl0
srfm smol
srad 0.0
swin 0.3
sdens 10.0
temp 298.15
gamma 0.105
calcenergy no
calcforce no
write pot dx complex
end
quit
|
$ apbs apbs-graphics.in
|
In what follows, we use the Tcl scripts my_functions and all.vmd to visualize the electrostatic potential data using VMD. The my_function Tcl script contains functions to load all 3 data sets; the user simply needs to type (at the system prompt):
$ vmd -e all.vmd
|
The next command loads the apo PKA electrostatic potential and is run from within VMD:
vmd > load_apo
|
Finally, the complex can be loaded by typing, from within VMD:
vmd > load_complex
|