

The X-Score program is developed by Dr. Renxiao Wang in Dr. Shaomeng Wang's group in the Department of Internal Medicine, University of Michigan Medical School. Here the prefix "X" indicates that it is part of the computer-aided drug design toolkit, X-Tool, which is being developed by Dr. Renxiao Wang. X-Score was formerly known as X-CScore.
X-Score estimates binding free energies for protein-ligand complexes with known 3D structures. Three empirical scoring functions have been implemented in this program, which are named as HPScore, HMScore, and HSScore:
HPScore = 2.783 - 0.001*(VDW) + 0.412*(H-Bond) + 0.037*(Hydrophobic Pair) - 0.100*(Rotor)
HMScore = 3.099 - 0.002*(VDW) + 0.311*(H-Bond) + 0.602*(Hydrophobic Match) - 0.169*(Rotor)
HSScore = 2.686 - 0.002*(VDW) + 0.307*(H-Bond) + (0.007)*(Hydrophobic surface) - 0.159*(Rotor)
By default, the final results: X-Score = (HPScore + HMScore + HSScore) / 3.
All these scoring functions give absolute dissociate constant of the given protein-ligand complex in negative logarithm (pKd). They are calibrated by a diverse set of 200 protein-ligand complexes and they are able to reproduce the binding affinities of the entire calibration set with an average accuracy of 1.5 pKd units in dissociation constant (~ 2 kcal/mol in binding free energy).
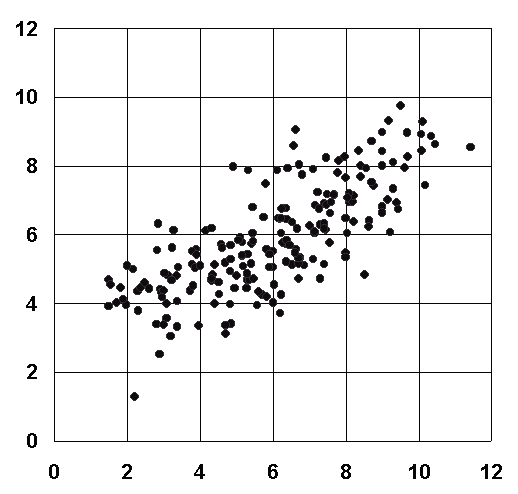
Correlation
between the experimental pKd values (X axis) and
the calculated pKd values (Y axis) by X-Score of 200 protein-ligand
complexes.
X-Score is typically applied in combination with a molecular docking program, such as DOCK, AutoDock, FlexX or GOLD, in structure-base drug design studies. The molecular docking program will provide the binding model of the molecules of interests to the given target protein. Then, X-Score can be applied to give more accurate estimation of the binding free energies of these molecules and re-rank them accordingly..
The correct way to refer this program is either X-Tool/Score or the shorter form, X-Score. The following reference is required to be cited in any resulting publication.
[Content] [Introduction] [Download] [Installation] [Usage] [Trouble Shooting]